Redun¶
Here, we’ll see how to track redun workflow runs with LaminDB.
Note
This use case is based on github.com/ricomnl/bioinformatics-pipeline-tutorial.
Amend the workflow¶
Here is how to instrument a redun workflow for tracking with lamindb:
Add
ln.track()(see on GitHub)Register desired output files or folders by creating artifacts for them (see on GitHub):
ln.Artifact(output_path, key="data/results.tar.gz").save()
Add a
finish()task that callsln.finish()(see on GitHub)Optionally cache/stage input files (see on GitHub)
Why not use @ln.flow() for main()?
Because main() in redun is typically a scheduler/executor task rather than a task that performs the actual computation. ln.flow() would hence measure the execution time of scheduling not an actual compute run.
If one wanted to use @ln.flow() it’s advisable to wrap the scheduler:
@ln.flow()
def run_pipeline(...):
scheduler = Scheduler()
result = scheduler.run(main(...)) # run the main task
ln.Artifact(result.path, key="data/results.tgz").save()
return result
Run redun¶
Let’s see what the input files are:
!ls ./fasta
Show code cell output
KLF4.fasta MYC.fasta PO5F1.fasta SOX2.fasta
Create a lamindb test instance:
# pip install lamindb redun git+http://github.com/laminlabs/redun-lamin-fasta
!lamin init --storage ./test-redun-lamin
→ initialized lamindb: testuser1/test-redun-lamin
Register each input file individually as an artifact:
import lamindb as ln
import json
ln.Artifact.from_dir("./fasta").save()
Show code cell output
→ connected lamindb: testuser1/test-redun-lamin
! folder is outside existing storage location, will copy files from ./fasta to /home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/fasta
! no run & transform got linked, call `ln.track()` & re-run
! no run & transform got linked, call `ln.track()` & re-run
! no run & transform got linked, call `ln.track()` & re-run
! no run & transform got linked, call `ln.track()` & re-run
SQLRecordList([Artifact(uid='VAmAHxJB3geN6W5c0000', version_tag=None, is_latest=True, key='fasta/SOX2.fasta', description=None, suffix='.fasta', kind=None, otype=None, size=414, hash='C5q_yaFXGk4SAEpfdqBwnQ', n_files=None, n_observations=None, branch_id=1, space_id=1, storage_id=3, run_id=None, schema_id=None, created_by_id=3, created_at=2026-01-27 14:09:42 UTC, is_locked=False),
Artifact(uid='KhTGrTaUZurrtiBY0000', version_tag=None, is_latest=True, key='fasta/MYC.fasta', description=None, suffix='.fasta', kind=None, otype=None, size=536, hash='WGbEtzPw-3bQEGcngO_pHQ', n_files=None, n_observations=None, branch_id=1, space_id=1, storage_id=3, run_id=None, schema_id=None, created_by_id=3, created_at=2026-01-27 14:09:42 UTC, is_locked=False),
Artifact(uid='mAzxFWtj4rC4aBpw0000', version_tag=None, is_latest=True, key='fasta/PO5F1.fasta', description=None, suffix='.fasta', kind=None, otype=None, size=477, hash='-7iJgveFO9ia0wE1bqVu6g', n_files=None, n_observations=None, branch_id=1, space_id=1, storage_id=3, run_id=None, schema_id=None, created_by_id=3, created_at=2026-01-27 14:09:42 UTC, is_locked=False),
Artifact(uid='IsrJucUuFnYiyXU30000', version_tag=None, is_latest=True, key='fasta/KLF4.fasta', description=None, suffix='.fasta', kind=None, otype=None, size=609, hash='LyuoYkWs4SgYcH7P7JLJtA', n_files=None, n_observations=None, branch_id=1, space_id=1, storage_id=3, run_id=None, schema_id=None, created_by_id=3, created_at=2026-01-27 14:09:42 UTC, is_locked=False)])
Run the redun workflow:
!redun run workflow.py main --input-dir ./fasta --tag run=test-run 1> run_logs.txt 2>run_logs.txt
Inspect the logs:
!cat run_logs.txt
Show code cell output
→ connected lamindb: testuser1/test-redun-lamin
→ script invoked with: run workflow.py main --input-dir ./fasta --tag run=test-run
→ created Transform('mwXpIb5MM9ju0000', key='workflow.py'), started new Run('3WFYYYxLncitfNLs') at 2026-01-27 14:09:53 UTC
• recommendation: to identify the script across renames, pass the uid: ln.track("mwXpIb5MM9ju")
File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/results.tgz, hash=d262b169)
_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/VAmAHxJB3geN6W5c0000.fasta, hash=a2bb69fa), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job 91656526: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/KhTGrTaUZurrtiBY0000.fasta, hash=e5dee22d), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job 3db612e5: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/mAzxFWtj4rC4aBpw0000.fasta, hash=7836a098), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job 083caf2b: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/IsrJucUuFnYiyXU30000.fasta, hash=97624216), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job c747e1dd: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/VAmAHxJB3geN6W5c0000.fasta, hash=a2bb69fa), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/VAmAHxJB3geN6W5c0000.peptides.txt, hash=a0430d03), amino_acid='C') on default
[redun] Run Job a29b67e6: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/KhTGrTaUZurrtiBY0000.fasta, hash=e5dee22d), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/KhTGrTaUZurrtiBY0000.peptides.txt, hash=9a8c6c07), amino_acid='C') on default
[redun] Run Job 27c74a06: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/mAzxFWtj4rC4aBpw0000.fasta, hash=7836a098), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/mAzxFWtj4rC4aBpw0000.peptides.txt, hash=e77e9352), amino_acid='C') on default
[redun] Run Job e10cacc3: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/IsrJucUuFnYiyXU30000.fasta, hash=97624216), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/IsrJucUuFnYiyXU30000.peptides.txt, hash=0e840446), amino_acid='C') on default
[redun] Run Job dc14db6a: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/VAmAHxJB3geN6W5c0000.count.tsv, hash=e4847ee0)) on default
[redun] Run Job 3a23ea75: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/KhTGrTaUZurrtiBY0000.count.tsv, hash=db248942)) on default
[redun] Run Job cf944aef: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/mAzxFWtj4rC4aBpw0000.count.tsv, hash=0d22a43a)) on default
[redun] Run Job 7c079b77: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/IsrJucUuFnYiyXU30000.count.tsv, hash=77d38a6e)) on default
[redun] Run Job c1a1051c: bioinformatics_pipeline_tutorial.lib.get_report_task(input_counts=[File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/VAmAHxJB3geN6W5c0000.count.tsv, hash=e4847ee0), File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-l...) on default
[redun] Run Job fb8723db: bioinformatics_pipeline_tutorial.lib.archive_results_task(inputs_plots=[File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/VAmAHxJB3geN6W5c0000.plot.png, hash=5836a8d8), File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-la..., input_report=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/protein_report.tsv, hash=86de8988)) on default
[redun] Run Job 5fd67f5e: redun_lamin_fasta.finish(results_archive=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/results.tgz, hash=d262b169)) on default
[redun]
[redun] | JOB STATUS 2026/01/27 14:09:58
[redun] | TASK PENDING RUNNING FAILED CACHED DONE TOTAL
[redun] |
[redun] | ALL 0 0 0 0 16 16
[redun] | bioinformatics_pipeline_tutorial.lib.archive_results_task 0 0 0 0 1 1
[redun] | bioinformatics_pipeline_tutorial.lib.count_amino_acids_task 0 0 0 0 4 4
[redun] | bioinformatics_pipeline_tutorial.lib.digest_protein_task 0 0 0 0 4 4
[redun] | bioinformatics_pipeline_tutorial.lib.get_report_task 0 0 0 0 1 1
[redun] | bioinformatics_pipeline_tutorial.lib.plot_count_task 0 0 0 0 4 4
[redun] | redun_lamin_fasta.finish 0 0 0 0 1 1
[redun] | redun_lamin_fasta.main 0 0 0 0 1 1
[redun]
[redun]
[redun] Execution duration: 5.55 seconds
View data lineage:
artifact = ln.Artifact.get(key="data/results.tgz")
artifact.view_lineage()
Show code cell output
artifact.transform.describe()
Transform: workflow.py (0000) ├── uid: mwXpIb5MM9ju0000 │ hash: zjZMEeofUMp3FV6fxbgjYg type: script │ branch: main space: all │ created_at: 2026-01-27 14:09:53 UTC created_by: testuser1 └── source_code: │ """workflow.py.""" │ │ # This code is based on a copy from https://github.com/ricomnl/bioinformatics-pi … │ # Copyright Rico Meinl 2022 │ from enum import Enum │ │ import lamindb as ln │ from redun import File, task │ │ import redun_lamin_fasta │ from redun_lamin_fasta.lib import ( │ archive_results_task, │ count_amino_acids_task, │ digest_protein_task, │ get_report_task, │ plot_count_task, │ ) │ │ redun_namespace = redun_lamin_fasta.__name__ │ │ │ class Executor(str, Enum): │ default = "default" │ process = "process" │ batch = "batch" │ batch_debug = "batch_debug" │ │ │ @task() │ def finish(results_archive: File) -> File: │ …
artifact.run.describe()
Run: 3WFYYYx (workflow.py) ├── uid: 3WFYYYxLncitfNLs transform: workflow.py (0000) │ started_at: 2026-01-27 14:09:53 UTC finished_at: 2026-01-27 14:09:58 UTC │ status: completed │ branch: main space: all │ created_at: 2026-01-27 14:09:53 UTC created_by: testuser1 ├── cli_args: │ │ run workflow.py main --input-dir ./fasta --tag run=test-run ├── report: ktTXjsD │ │ → connected lamindb: testuser1/test-redun-lamin │ │ → created Transform('mwXpIb5MM9ju0000', key='workflow.py'), started new Run('3WF … │ │ • recommendation: to identify the script across renames, pass the uid: ln.track( … └── environment: XeG2Kb3 │ aiobotocore==2.26.0 │ aiohappyeyeballs==2.6.1 │ aiohttp==3.13.3 │ aioitertools==0.13.0 │ …
View transforms and runs in LaminHub¶
Explore it at lamin.ai/laminlabs/lamindata/transform/taasWKawCiNA.
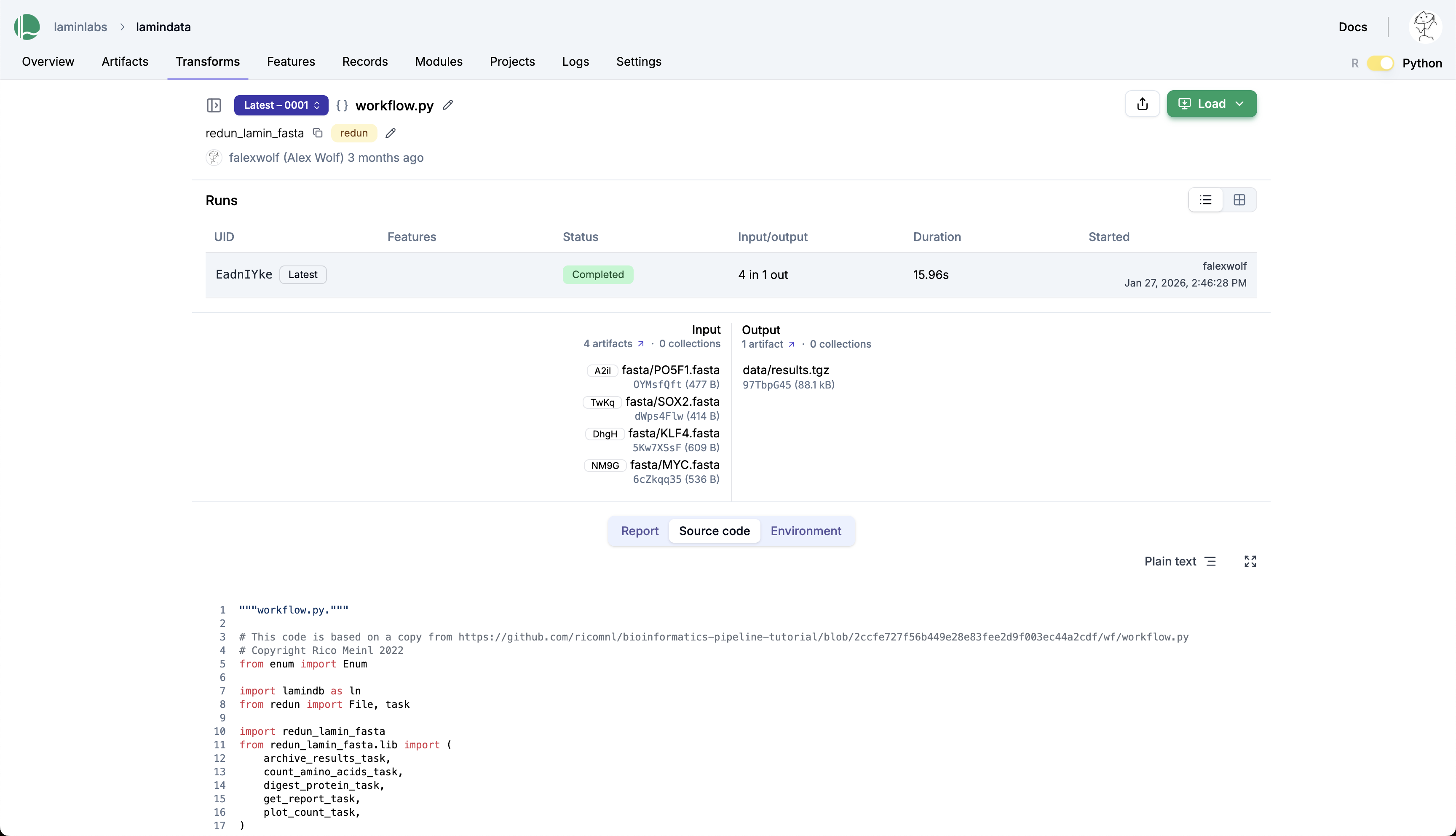
View the database content¶
ln.view()
Artifact
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | version_tag | is_latest | is_locked | created_at | branch_id | space_id | storage_id | run_id | schema_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||||
| 6 | 455oC3z76p1BaoN50000 | data/results.tgz | None | .tgz | None | None | 94326 | Q-3nswv28Izrox1EdzagtQ | None | None | None | True | False | 2026-01-27 14:09:58.663000+00:00 | 1 | 1 | 3 | 1.0 | None | 3 |
| 4 | IsrJucUuFnYiyXU30000 | fasta/KLF4.fasta | None | .fasta | None | None | 609 | LyuoYkWs4SgYcH7P7JLJtA | None | None | None | True | False | 2026-01-27 14:09:42.980000+00:00 | 1 | 1 | 3 | NaN | None | 3 |
| 3 | mAzxFWtj4rC4aBpw0000 | fasta/PO5F1.fasta | None | .fasta | None | None | 477 | -7iJgveFO9ia0wE1bqVu6g | None | None | None | True | False | 2026-01-27 14:09:42.980000+00:00 | 1 | 1 | 3 | NaN | None | 3 |
| 2 | KhTGrTaUZurrtiBY0000 | fasta/MYC.fasta | None | .fasta | None | None | 536 | WGbEtzPw-3bQEGcngO_pHQ | None | None | None | True | False | 2026-01-27 14:09:42.979000+00:00 | 1 | 1 | 3 | NaN | None | 3 |
| 1 | VAmAHxJB3geN6W5c0000 | fasta/SOX2.fasta | None | .fasta | None | None | 414 | C5q_yaFXGk4SAEpfdqBwnQ | None | None | None | True | False | 2026-01-27 14:09:42.978000+00:00 | 1 | 1 | 3 | NaN | None | 3 |
Run
| uid | name | entrypoint | started_at | finished_at | params | reference | reference_type | cli_args | is_locked | created_at | branch_id | space_id | transform_id | report_id | environment_id | created_by_id | initiated_by_run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 1 | 3WFYYYxLncitfNLs | None | None | 2026-01-27 14:09:53.384607+00:00 | 2026-01-27 14:09:58.692803+00:00 | None | None | None | run workflow.py main --input-dir ./fasta --tag... | False | 2026-01-27 14:09:53.385000+00:00 | 1 | 1 | 1 | 7 | 5 | 3 | None |
Storage
| uid | root | description | type | region | instance_uid | is_locked | created_at | branch_id | space_id | created_by_id | run_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||
| 3 | 0X75sxUGrYCg | /home/runner/work/redun-lamin/redun-lamin/docs... | None | local | None | iQlBPgD8uaqR | False | 2026-01-27 14:09:40.142000+00:00 | 1 | 1 | 3 | None |
Transform
| uid | key | description | kind | source_code | hash | reference | reference_type | version_tag | is_latest | is_locked | created_at | branch_id | space_id | environment_id | created_by_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||
| 1 | mwXpIb5MM9ju0000 | workflow.py | None | script | """workflow.py."""\n\n# This code is based on ... | zjZMEeofUMp3FV6fxbgjYg | None | None | None | True | False | 2026-01-27 14:09:53.382000+00:00 | 1 | 1 | None | 3 |
Appendix¶
Map the redun execution id¶
If we want to be able to query LaminDB for redun execution ID, this here is a way to get it:
# export the run information from redun
!redun log --exec --exec-tag run=test-run --format json --no-pager > redun_exec.json
# load the redun execution id from the JSON and store it in the LaminDB run record
with open("redun_exec.json") as file:
redun_exec = json.loads(file.readline())
artifact.run.reference = redun_exec["id"]
artifact.run.reference_type = "redun_id"
artifact.run.save()
Run(uid='3WFYYYxLncitfNLs', name=None, entrypoint=None, started_at=2026-01-27 14:09:53 UTC, finished_at=2026-01-27 14:09:58 UTC, params=None, reference='1311fa4f-ccd1-42e8-8106-02690df824fc', reference_type='redun_id', cli_args='run workflow.py main --input-dir ./fasta --tag run=test-run', branch_id=1, space_id=1, transform_id=1, report_id=7, environment_id=5, created_by_id=3, initiated_by_run_id=None, created_at=2026-01-27 14:09:53 UTC, is_locked=False)
Map the redun run report¶
While lamindb auto-tracks the logs of the main python process you might also want to link the dedicated redun logs:
report = ln.Artifact(
"run_logs.txt",
description=f"Redun run report of {redun_exec['id']}",
run=False,
kind="__lamindb_run__", # mark as auxiliary artifact for the run
).save()
artifact.run.report = report
artifact.run.save()
Run(uid='3WFYYYxLncitfNLs', name=None, entrypoint=None, started_at=2026-01-27 14:09:53 UTC, finished_at=2026-01-27 14:09:58 UTC, params=None, reference='1311fa4f-ccd1-42e8-8106-02690df824fc', reference_type='redun_id', cli_args='run workflow.py main --input-dir ./fasta --tag run=test-run', branch_id=1, space_id=1, transform_id=1, report_id=8, environment_id=5, created_by_id=3, initiated_by_run_id=None, created_at=2026-01-27 14:09:53 UTC, is_locked=False)
Delete the test instance:
Show code cell content
!rm -rf test-redun-lamin
!lamin delete --force test-redun-lamin

